


Now faculty member at University of Alaska, Fairbanks.
current graduate student
 |
 |
 |
| Eric Collins Now faculty member at University of Alaska, Fairbanks. |
Seyed Ali Reza Zamani Dahaj current graduate student |
Paul Higgs |
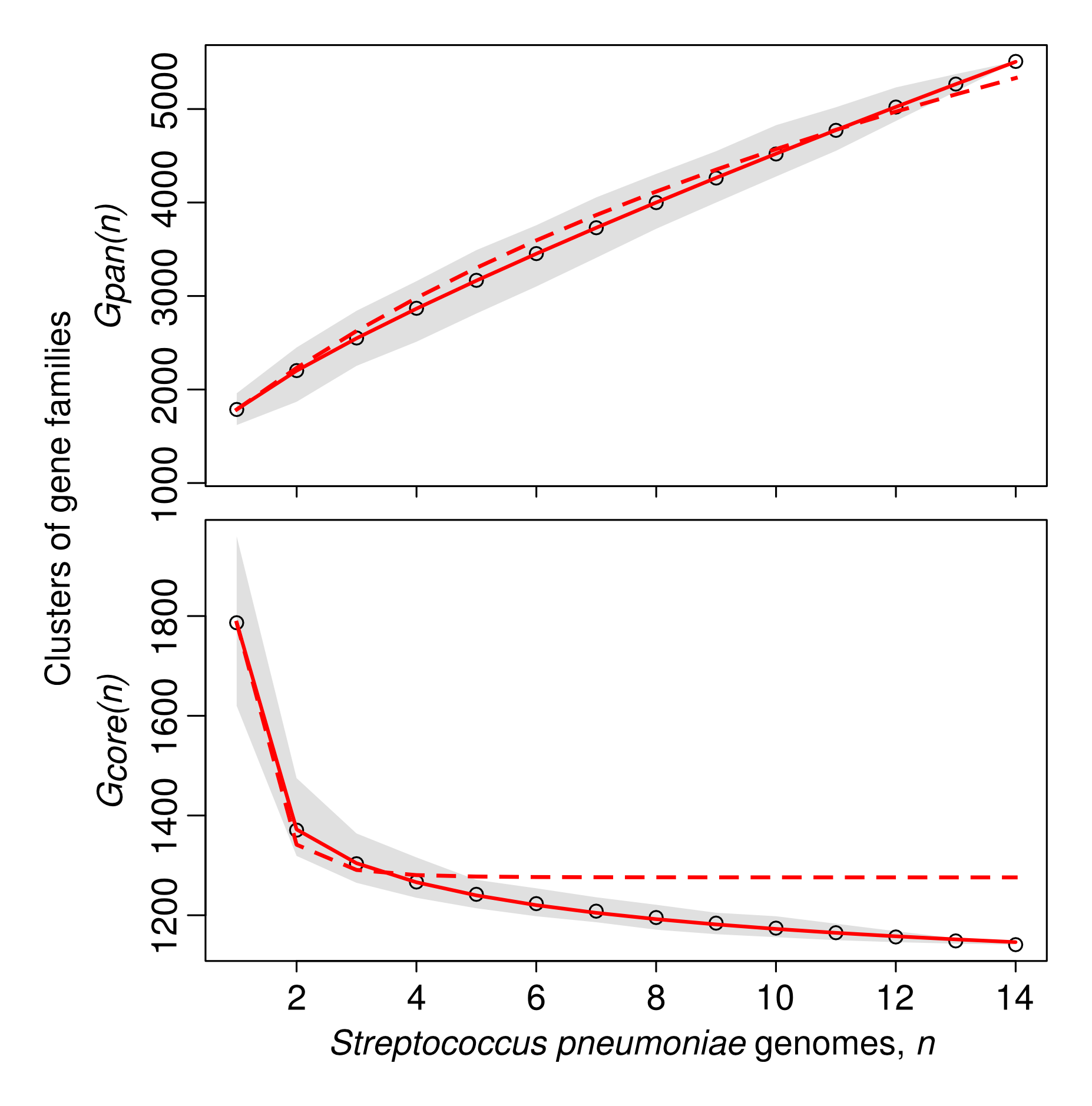
Bacteria gain and lose genes surprisingly rapidly. Even genomes that belong nominally to the same species do not have the same set of genes. In this example (from Collins and Higgs [1]) all the genes from the genomes of 14 strains of Streptococcus pneumoniae were clustered according to sequence similarity.
The Pangenome is the set of genes found at least once in a set of genomes. Gpan(n) is the number of gene families found after n genomes are included. There are about 1800 genes in each genome, but a total of more than 5000 genes in the pangenome of 14 strains. The pangenome size continues to increase each time a new genome is added because each new genome contains genes not seen previously. The Core Genome is the set of genes found on every genome in the set. Gcore(n) falls to only 1200 after including all 14 genomes.
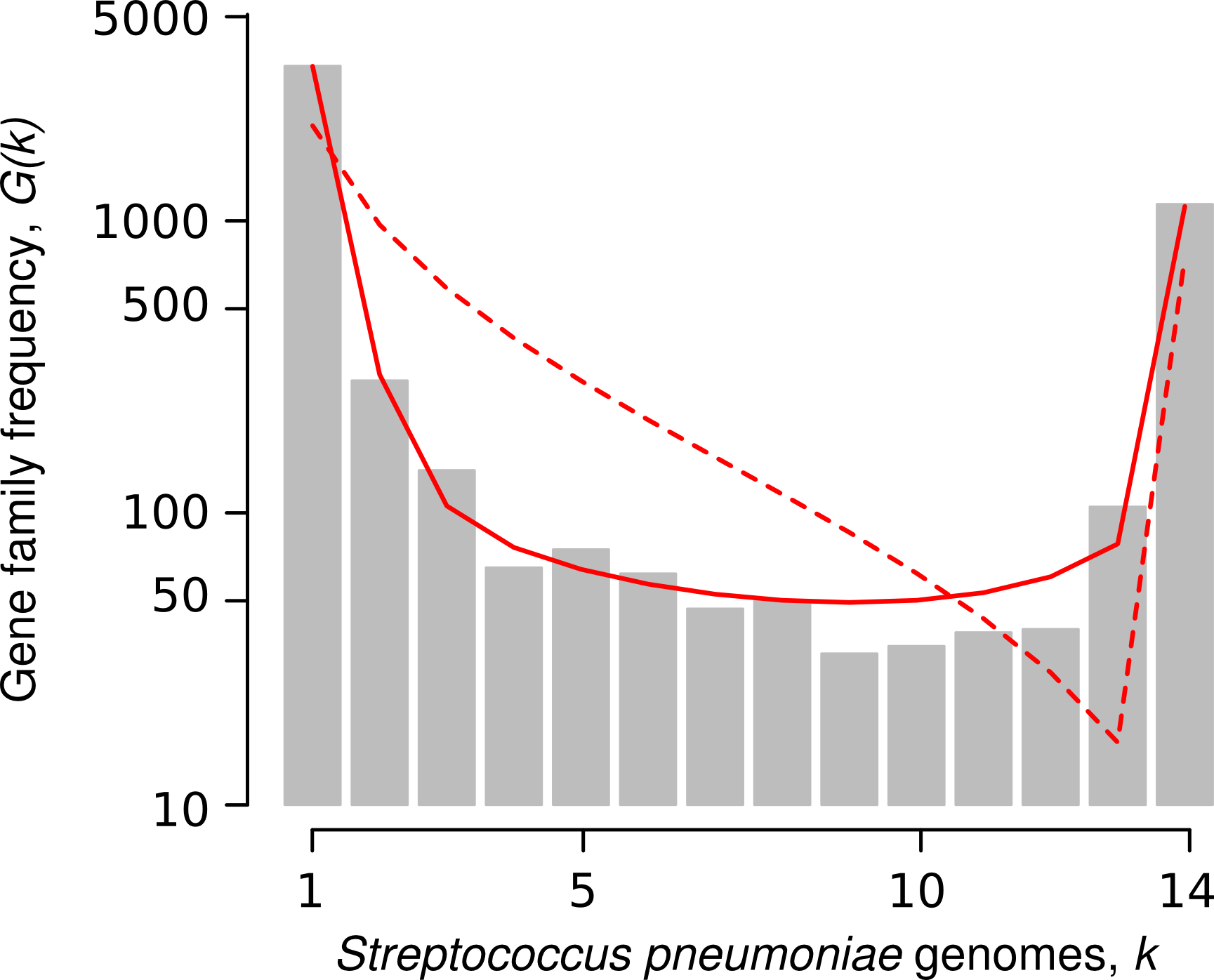
The gene frequency spectrum, G(k), is the number of gene families that are present in k genomes. This has a U-shape distribution with some core genes found in all 14 genomes and a large number of genes found in only 1 or 2 genomes. These latter genes have probably originated very recently, either by sequence evolution within the genome or Horizontal Gene Transfer from outside the group.
The phylogenetic tree of bacteria from the class Bacilli is shown below, calculated using conserved genes found in all Bacilli. The Streptococcus genomes considered above are a very closely related subgroup within this tree.
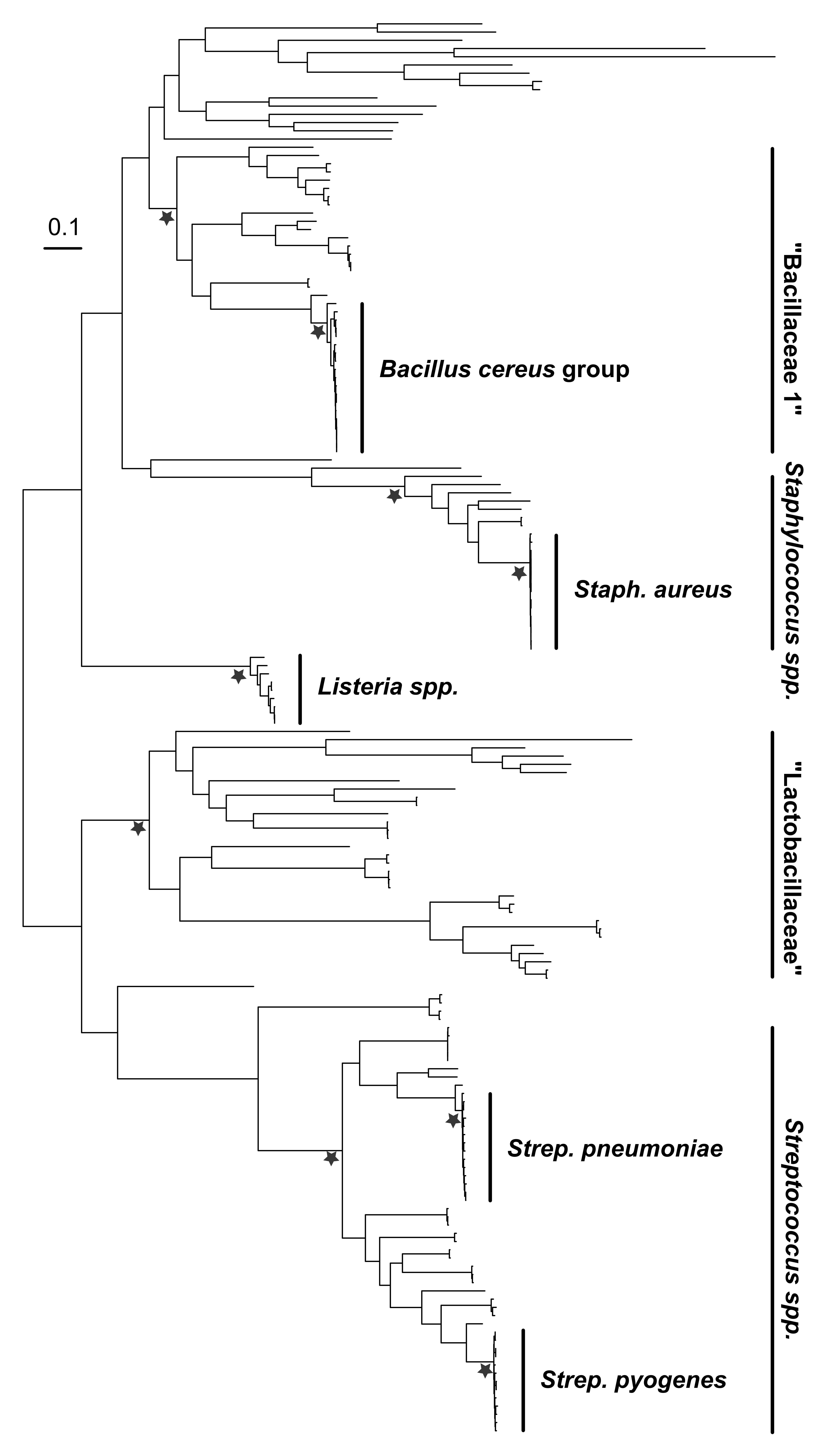
The fact that many genes are inserted and deleted on the timescale of divergence between the Strep. pneumoniae strains suggests that horizontal transfer can be very rapid. It is sometimes claimed that horizontal transfer is so frequent that it is not possible to draw phylogenetic trees for bacteria. However, there are substantial numbers of genes that are conserved over long periods of time, and reliable trees can be inferred from these genes. In our view, a strong signal of treelike evolution (vertical inheritance of genes) remains, despite the presence of substantial horizontal transfer.