

Now a Postdoc at NCBI
Current graduate student
Current undergraduate student
| |
 |
 |
| Wenqi Ran Now a Postdoc at NCBI |
Arash Ahmadi Current graduate student |
Aamer Somani Current undergraduate student |
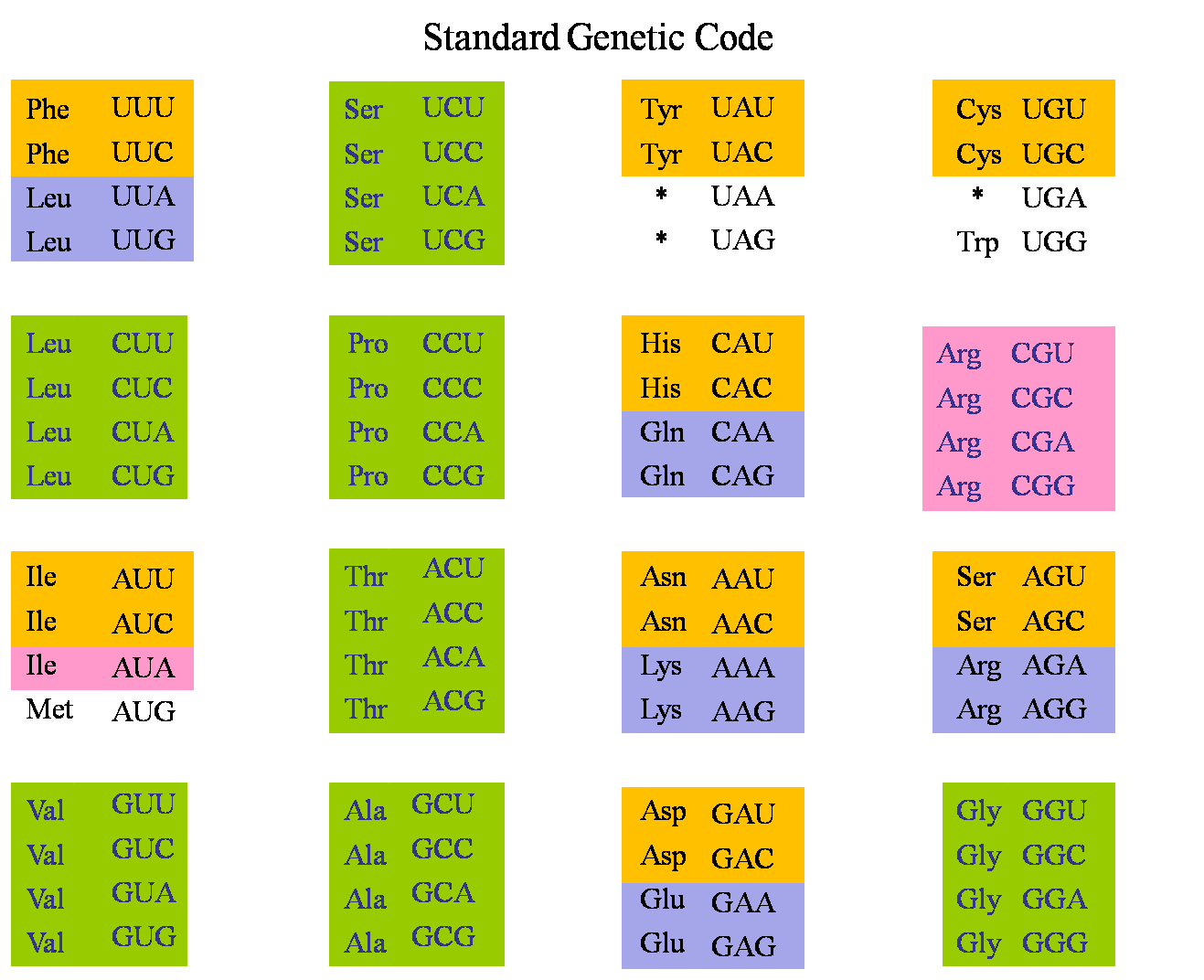
Most amino acids are encoded by more than one amino acid in the genetic code. Alternative codons for the same amino acid (synonymous codons) are not used with equal frequency. Codons that are more rapidly translated by the ribosome are used more frequently (particularly in highly expressed genes). This is known as selection for translational efficiency.
The choice of base at the synonymous third position of the codon is related to the base at the wobble position of the tRNA (the first anticodon base, or position 34 of the tRNA).
 |
 |
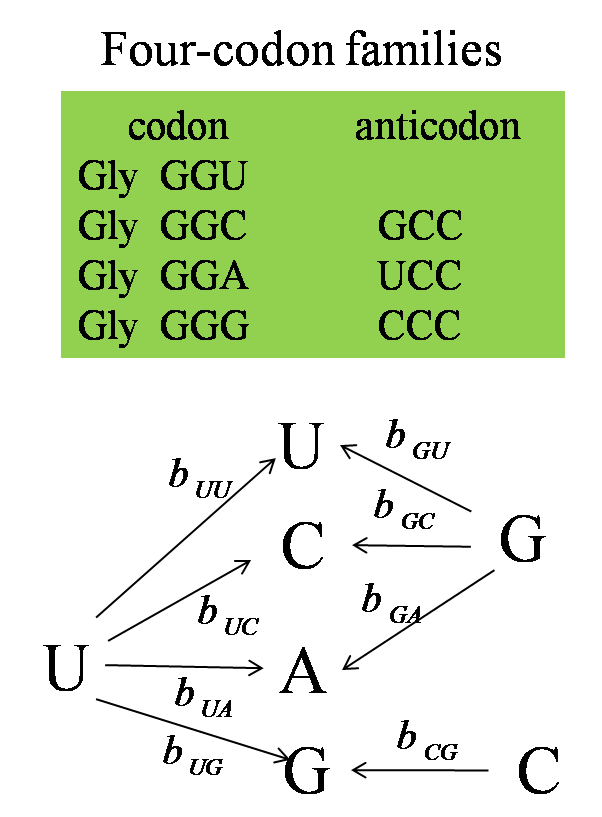 |
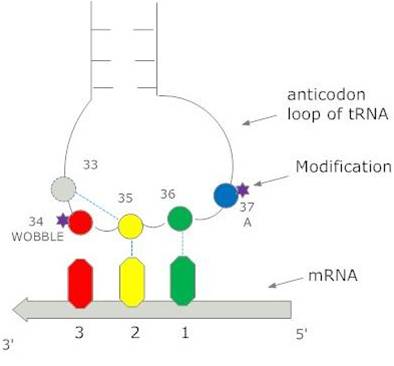
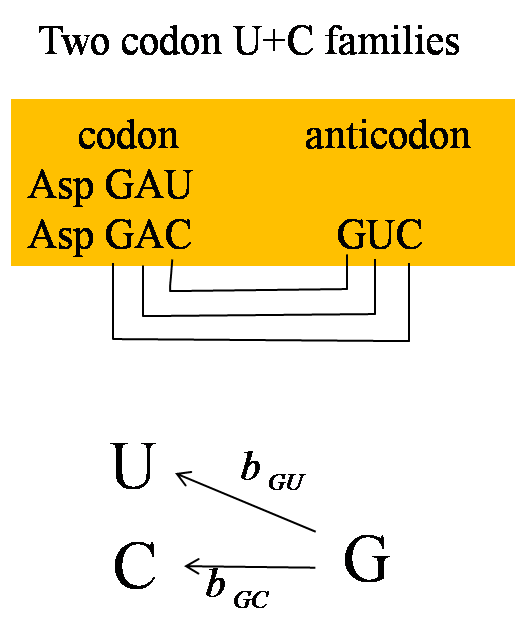
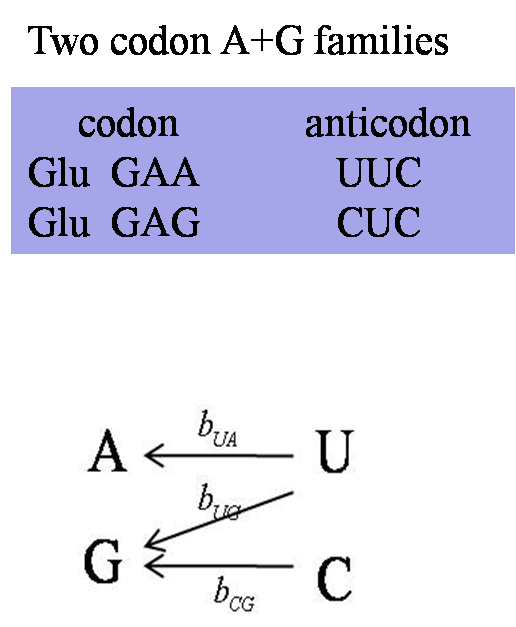
| In a two-codon U+C family, the wobble position is always a G and a single tRNA can translate both U and C codons. The C codon is preferred in almost all U+C families in almost all bacteria that we studied. Most likely, the ribosome recognizes the tRNA more rapidly when the third position base is a C than when it is a U. | In a two-codon A+G family, there is always a tRNA with a U at the wobble position, and there is sometimes an additional tRNA with a wobble C. The direction of selection on the codon usage depends on the relative numbers of these two tRNAs. | In a four-codon family, there is always a tRNA with a wobble U, and there can sometimes be tRNAs with wobble G and wobble C. The preferred codon is dependent on the relative numbers of these tRNAs. |
The wobble base of tRNAs is often modified to a non-standard base, and the presence of these base modifications has important effects on the codon-anticodon interaction, and hence the direction of selection on codon usage. Our published papers condsider these effects in detail in a large number of bacterial genomes.
 |
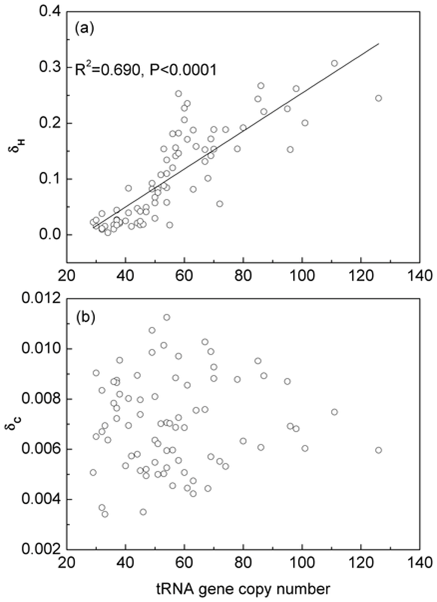 |
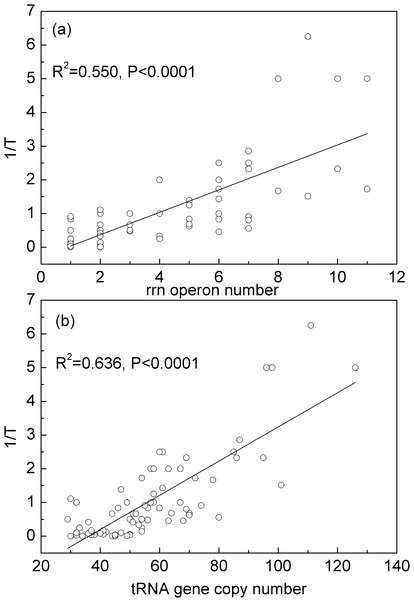
| The minimum cell division time, T, for bacteria varies between ~15 minutes and several days, depending on the niche/lifestyle that the bacteria adopt. Protein synthesis is a limiting step determining the rate of cell growth and division. The maximum growth rate, 1/T, is strongly correlated with both the number of ribosomal RNAs in the genome and the number of tRNAs. Duplication of rRNA genes allows faster ribosome synthesis and increased number of ribosomes. Duplication of tRNA genes allows faster translation by each ribosome. | Codon bias is strongest in organisms that have more tRNAs in the genome (i.e. the fast-growing organisms). δH measures the difference in codon frequencies between high-expression and low-expression genes. This is thought to be due to selection for speed/efficiency. δC measures the difference in codon frequencies between conserved and variable sites in genes. This is thought to arise from selection for translational accuracy. Our results show that selection for accuracy seems to be a weak effect and is not correlated with tRNA gene-number or growth rate, whereas selection for speed is a much stronger effect that is clearly correlated with both tRNA number and growth rate. |
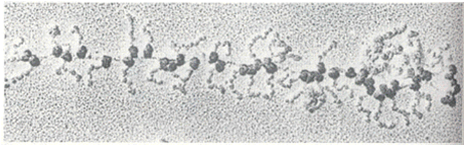
Often, more than one ribosome is translating a given mRNA at the same time, as shown in this image. If the density of ribosomes is high, they can potentially interfere with one another. The presence of slow codons within a sequence can cause a bottlenecks and a ribosomal traffic jam.
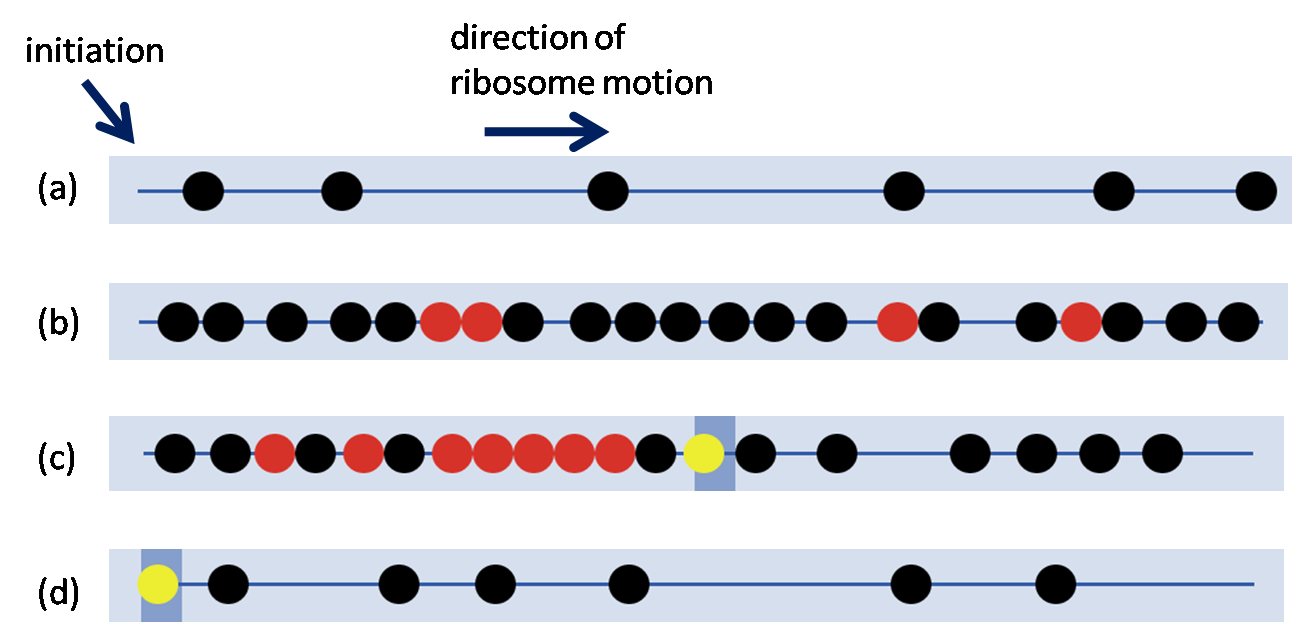
These figures show simulations of a model of particles moving along a one-dimensional track, representing the dynamics of ribosomes.