Publications
The genetic code is the set of assignments between the 64 codons in DNA and the 20 amino acids. The code is almost universal and arose prior to the latest common ancestor of all current life. However, there are many cases of specific groups of organisms or organelles where the code has changed due to the reassignment of one or more codons to a new amino acid. We are studying the way these changes occur.
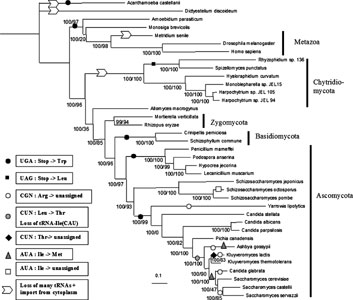
The above figure shows the positions of codon reassignments in mitochondrial genomes of fungi. Better images of the phylogenies of all eukaryotic groups showing the positions of all known codon reassignments in mitochondrial genomes are available here.
We developed a model that we call the Gain-Loss framework [1] that that incorporates four different scenarios for codon reassignment. Two of these (the codon disappearance mechanism, and the ambiguous intermediate mechanism) are found in the literature, and the other two (unassigned codon mechanism and compensatory change mechanism) were introduced by us. We have shown that all these mechanisms are possible within the same population genetics model, and using computer simulations, we studied the factors that influence the relative frequency of the different mechanisms. We were also able to determine the likely mechanisms of codon reassignments in mitochondrial genomes [2].
An important feature of the canonical genetic code is that it appears to be arranged so as to minimize the effect of mutations and errors in translation. Both these types of error are likely to cause a change in one base out of the three in the codon. Amino acid substitutions caused by single-base errors tend to be between amino acids of similar properties - hence the negative effects of the substitution are reduced. Statistical methods show that the canonical code is better than almost all random codes created by reshuffling the positions of the amino acids in the table of codons. We are now considering more carefully how this optimised state might have arisen. Most likely, there was a gradual development of the code by incorporation of additional amino acids.
It is likely that some amino acids were much more frequent than others at the time of the origin of life. Many possible methods of non-biological synthesis of amino acids have been proposed (atmospheric chemistry, deep sea vents, interstellar grains and meteorites). Although these theories are very diverse, there is a consensus that indicates which of the amino acids were frequent before life arose - about 10 of the 20 used in proteins today [3]. It seems likely that these were the first amino acids to be used in proteins, and that the remaining ones were added at a later date as the organisms evolved efficient pathways to synthesize them.