OGRe stands for Organelle Genome Retrieval. It is principally the work of Wenli Jia and Bin Tang at McMaster and Daniel Jameson at the University of Manchester. You can access the database online here: ogre.mcmaster.ca
OGRe is a relational database that contains over 1000 completely sequenced metazoan mitochondrial genome sequences. Using the database you can:
If you use information from OGRe, please cite:
The database now forms a resource for research on the evolution of codon usage, context-dependent mutation and genome rearrangements on mitochondrial genomes. See:
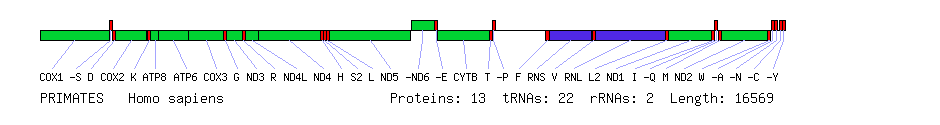
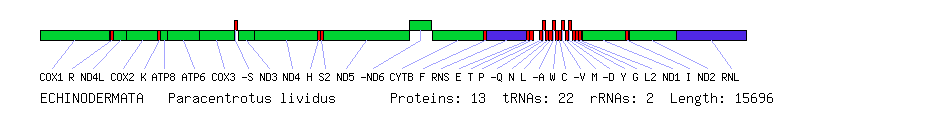
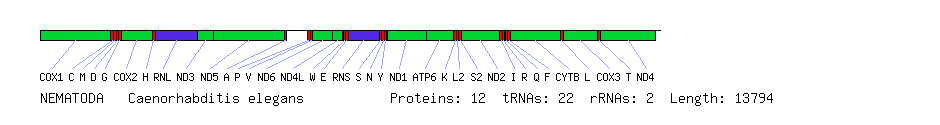
Comparison of human, sea urchin and nematode mitochondrial genomes. Protein genes are in green, rRNA genes in blue and tRNA genes in red. Boxes below the line represent genes on the forward strand (left to right) and boxes above the line represent genes on the reverse strand (right to left). All genes are on the same strand in the nematode. With the exception of the ATP8 gene, which is missing in the nematode, all three species have the same set of genes. The order differs greatly between the species, however.
 |
 |
 |
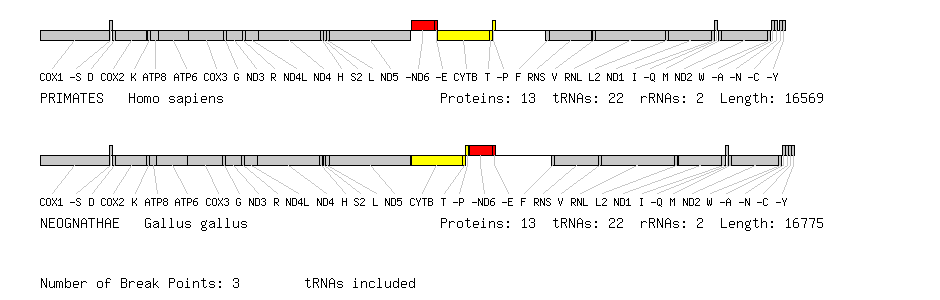
An example of a pairwise genome comparison produced by OGRe where tRNA genes are included. The blocks of genes coloured red and yellow have exchanged positions between human and chicken but all genes remain on the same strand.
 |
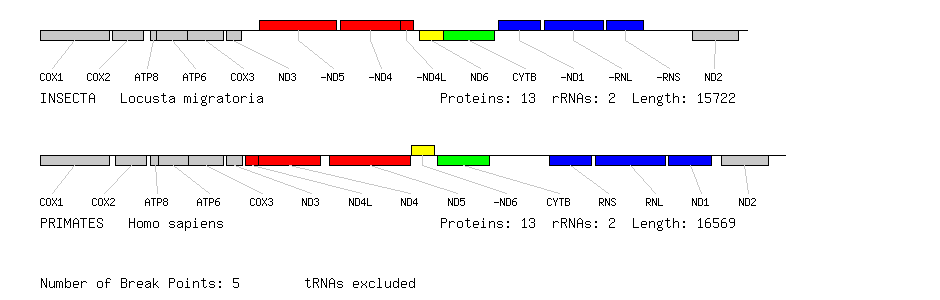
An example of a pairwise genome comparison produced by OGRe where tRNA genes are excluded. There are apparently three separate inversions of blocks of genes between human and locust (shown in red, yellow and blue). The grey and green gene blocks are unchanged.
 |