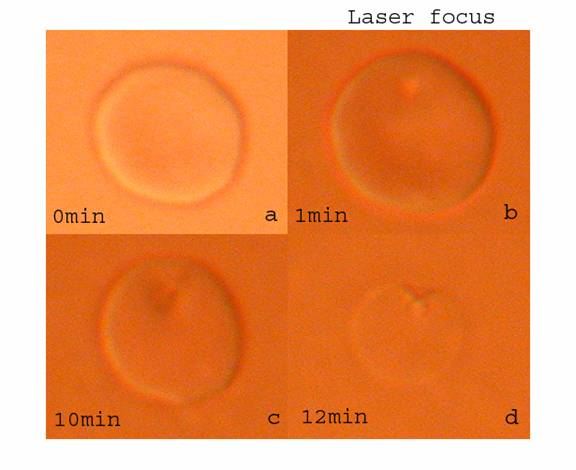
|
Daniel is investigating the diffusion of proteins in crowded environments using a model system
constituted of polymers and proteins.
-
Why study anomalous diffusion in crowded environments?
A cell depends on the diffusion of many of its proteins for
its proper functioning. Much study is being done on the function of
individual proteins, and how they are transported in the cell. Cells are full
of obstacles. Membranes, organelles, and other proteins obstruct diffusion.
To properly understand what is happening inside a cell, the possibility of
anomalous diffusion must be taken into account.
- What is anomalous diffusion?
An uninhibited particle in solution diffuses according to
Brownian motion, but in complicated environments the diffusion may become
anomalous. While the mean square displacement (MSD) of a particle following
normal diffusion is proportional to time: <r2(t)> ~ t,
anomalous diffusion is defined by: <r2(t)> ~ ta, with a<1. As a result, the apparent diffusion coefficient (D ~ <r2(t)>/t) is a function of time.
The diffusion rate of a particle motion obstructed by
obstacles depends on the size, number, and random positioning of the
obstacles. This makes an analytical determination of a general solution to
this problem enormously difficult, if not impossible. Instead, Monte
Carlo simulations and other numerical methods are used. The
result is of the form expected for anomalous diffusion: <r2(t)>
~ ta, with the anomalous
coefficient a depending on the
concentration, size, mobility and reactivity of the obstacles.
In case of anomalous diffusion, autocorrelation curves recorded with FCS
will have the form:
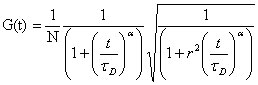
as opposed to:
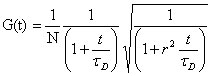 , ,
for normal 3-D diffusion.
- What is the model system in this study?
Our simple model uses a fluorescently labeled protein in solution with non-fluorescent obstacles
 (dextrans of different sizes). We can vary the concentration and size of the obstacles.
We can measure the diffusion coefficient and the anomalous exponent
characterizing the obstructed diffusion. We want to understand the diffusion
process in our model system, and to determine how the size and concentration
of obstacles influence this process. Our results will include a range of data
that should be useful for comparison with in vivo studies of anomalous
protein diffusion.
(dextrans of different sizes). We can vary the concentration and size of the obstacles.
We can measure the diffusion coefficient and the anomalous exponent
characterizing the obstructed diffusion. We want to understand the diffusion
process in our model system, and to determine how the size and concentration
of obstacles influence this process. Our results will include a range of data
that should be useful for comparison with in vivo studies of anomalous
protein diffusion.
References:
[1] Bunde, A.; Havlin, S.,
Eds. (1995) Fractals and Disordered Systems. 2nd edit., pp. 115- 175, Springer-Verlag, Berlin.
[2] Saxton, Micheal J. (1994) Anomalous Diffusion Due to Obstacles: A Monte Carlo Study. Biophysical Journal, 66:394-401.
[3] Wachsmuth, M., Waldeck, W. & Langowski, J. (2000) Anomalous diffusion
of fluorescent probes inside living cell nuclei investigated by
spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol.
298:677-689.
|




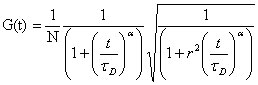
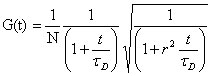 ,
,
 (dextrans of different sizes). We can vary the concentration and size of the obstacles.
We can measure the diffusion coefficient and the anomalous exponent
characterizing the obstructed diffusion. We want to understand the diffusion
process in our model system, and to determine how the size and concentration
of obstacles influence this process. Our results will include a range of data
that should be useful for comparison with in vivo studies of anomalous
protein diffusion.
(dextrans of different sizes). We can vary the concentration and size of the obstacles.
We can measure the diffusion coefficient and the anomalous exponent
characterizing the obstructed diffusion. We want to understand the diffusion
process in our model system, and to determine how the size and concentration
of obstacles influence this process. Our results will include a range of data
that should be useful for comparison with in vivo studies of anomalous
protein diffusion.